NEBUILDER HIFI DNA ASSEMBLY® Removal of 3´ end Mismatches
Script
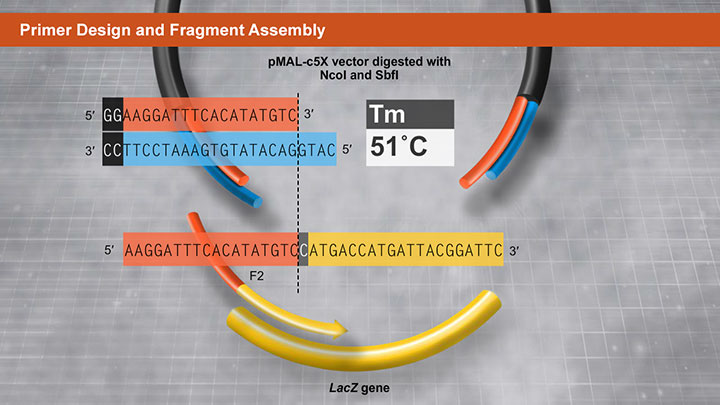
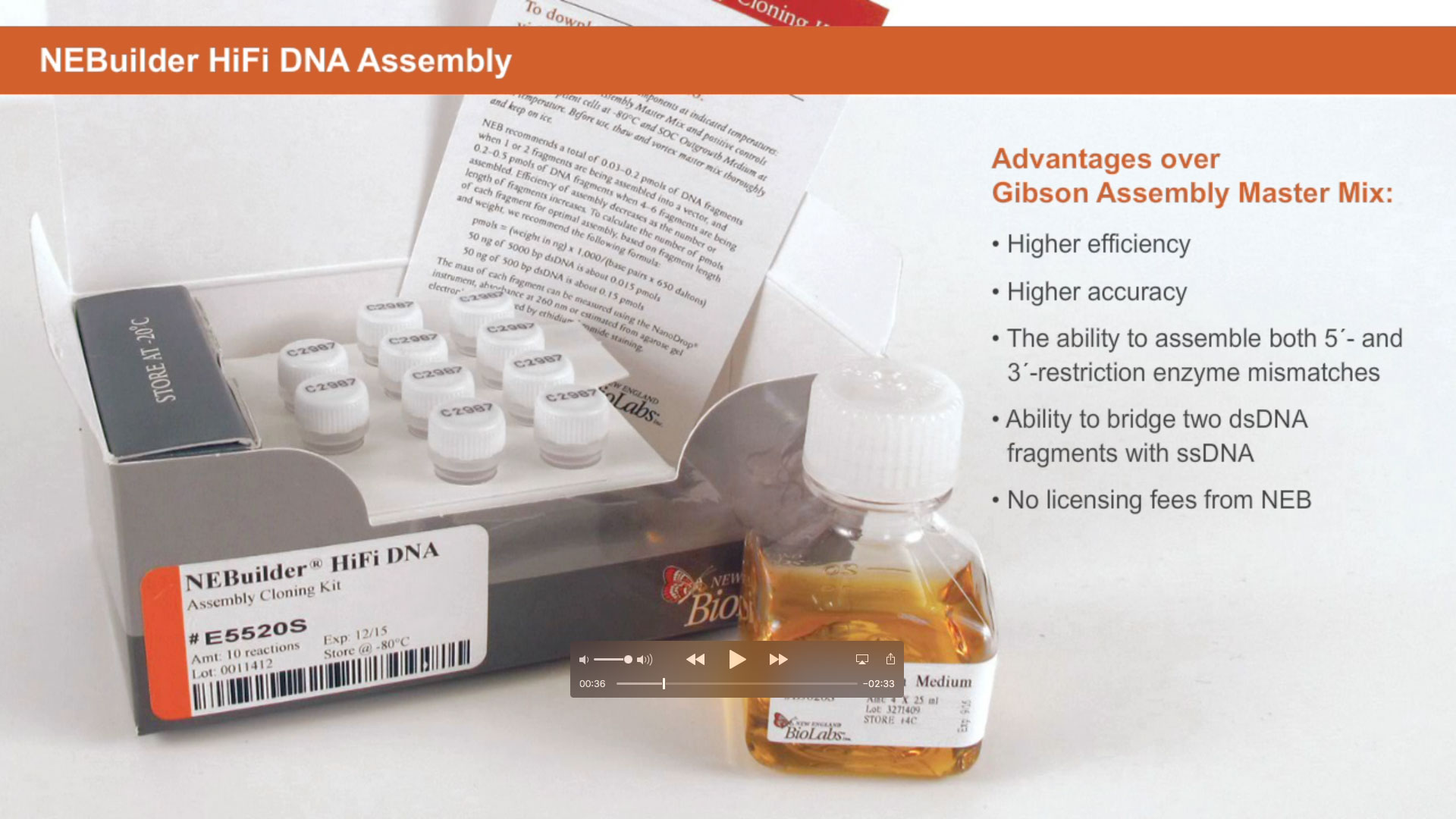
One of the unique features of the NEBuilder HiFi DNA Assembly Master Mix is the ability to remove 3-prime and 5-prime-end mismatch sequences upon fragment assembly.
During fragment assembly, T5 Exonuclease will remove the bases from the 5-prime end, generating a 3-prime overhang. Single-stranded 3-prime ends can then anneal…the DNA polymerase fills in the gap…and DNA ligase joins the adjacent fragments.
But, what happens if there are extra bases on the end of your 3-prime end that you do not want to incorporate into your assembly product? NEBuilder HiFi is capable of efficiently removing 3-prime-end mismatches, up to 10 base pairs from the 3-prime ends, so that they can be efficiently joined together.
In this example, we want to join a linearized vector and an insert at the 5-prime position of this restriction site. The insert is generated by PCR, and it has overlaps corresponding to the vector. When the overlaps anneal during the assembly process, there is a mismatch at the 3-prime end of the vector. The proofreading DNA polymerase removes the extra bases at the 3-prime end…the polymerase fills the gap…and DNA ligase seals the remaining nick.
Unlike other cloning methods, NEBuilder HiFi allows you to digest with restriction enzymes and conveniently assemble your DNA fragment at any position.
Related Videos
-
Primer Design and Fragment Assembly Using NEBuilder HiFi DNA Assembly® or Gibson Assembly® -
Introduction to NEBUILDER HIFI DNA ASSEMBLY® -

NEBUILDER® HiFi: the next generation of DNA Assembly

